ConSurf | Evolutionary conservation profiles of proteins, which analyze the sequence conservation of proteins and color the model by the result, it could be helpful if the proteins you studied are conserved on structure but vary on the sequence.
Analysis conservation
All you need to prepare is a protein model, you need the sequence at least if you don't have one, the structure can be predicted from it.
If you have a multiple sequence alignment(MSA) file, you can upload it, and then the sequence will be used to generate conservation analysis results.
Then your model will be colored from the results.
Customize color
There is no choice of color on the model in ConSurf, but you can do it with Pymol.
-
Download the result for Pymol.
-
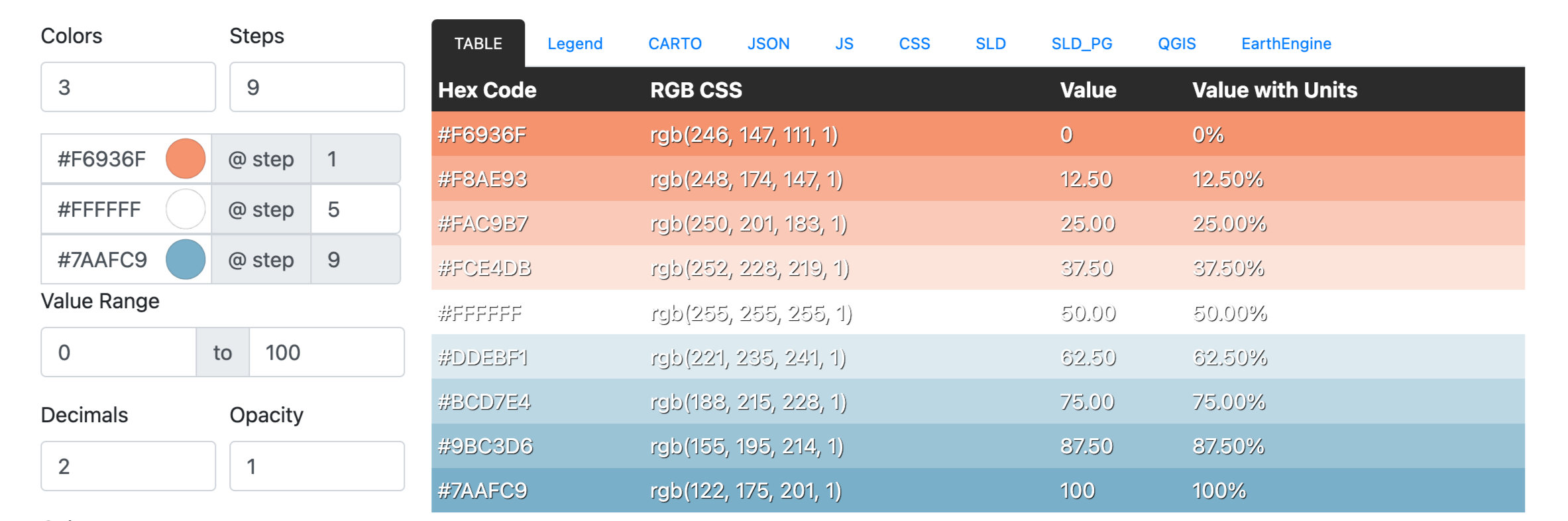
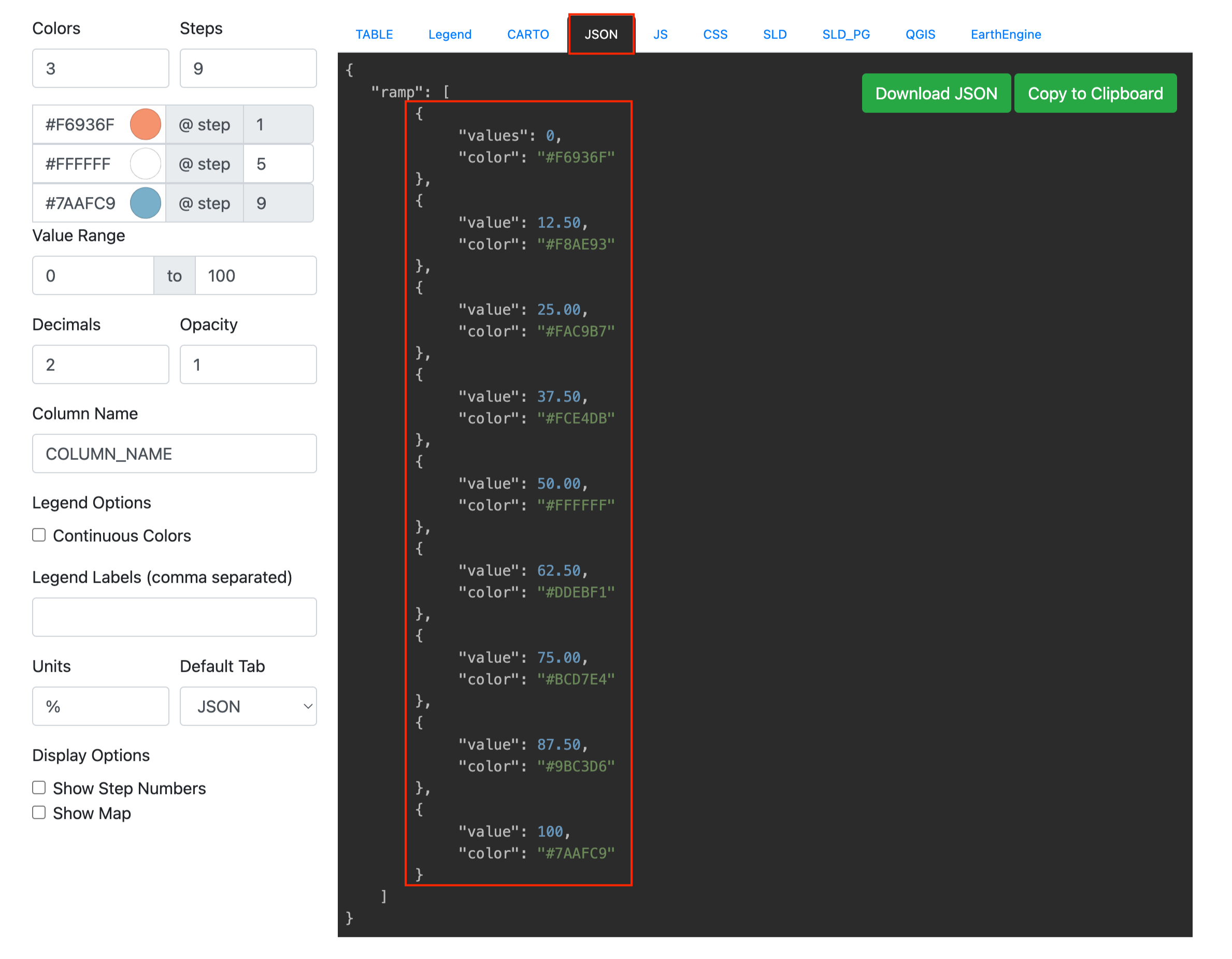
Generate a color ramp on RAMPGEN.COM, then copy the JSON format and replace the color in the script:
:::grid {cols=2,rows=1,gap=12,type=images}
::: -
Paste your color ramp into the script and save it as
set_colors.py:# This script was completed with the help of ChatGPT color_data = [ # Paste your color ramp here. ] # Define colors and set colors to corresponding objects for i, item in enumerate(color_data, start=1): hex_color = item['color'] # Convert HEX to RGB r, g, b = int(hex_color[1:3], 16), int(hex_color[3:5], 16), int(hex_color[5:], 16) rgb = (r/255.0, g/255.0, b/255.0) # Set the color color_name = f"color_{i}" pymol.cmd.set_color(color_name, rgb) # Apply color to objects object_name = f"all_group_{i}" pymol.cmd.color(color_name, object_name) # Ensure that views are updated pymol.cmd.rebuild() -
Your script should look like this:
color_data = [ { "values": 0, "color": "#F6936F" }, { "value": 12.50, "color": "#F8AE93" }, { "value": 25.00, "color": "#FAC9B7" }, { "value": 37.50, "color": "#FCE4DB" }, { "value": 50.00, "color": "#FFFFFF" }, { "value": 62.50, "color": "#DDEBF1" }, { "value": 75.00, "color": "#BCD7E4" }, { "value": 87.50, "color": "#9BC3D6" }, { "value": 100, "color": "#7AAFC9" } ] # Define colors and set colors to corresponding objects for i, item in enumerate(color_data, start=1): hex_color = item['color'] # Convert HEX to RGB r, g, b = int(hex_color[1:3], 16), int(hex_color[3:5], 16), int(hex_color[5:], 16) rgb = (r/255.0, g/255.0, b/255.0) # Set the color color_name = f"color_{i}" pymol.cmd.set_color(color_name, rgb) # Apply color to objects object_name = f"all_group_{i}" pymol.cmd.color(color_name, object_name) # Ensure that views are updated pymol.cmd.rebuild() -
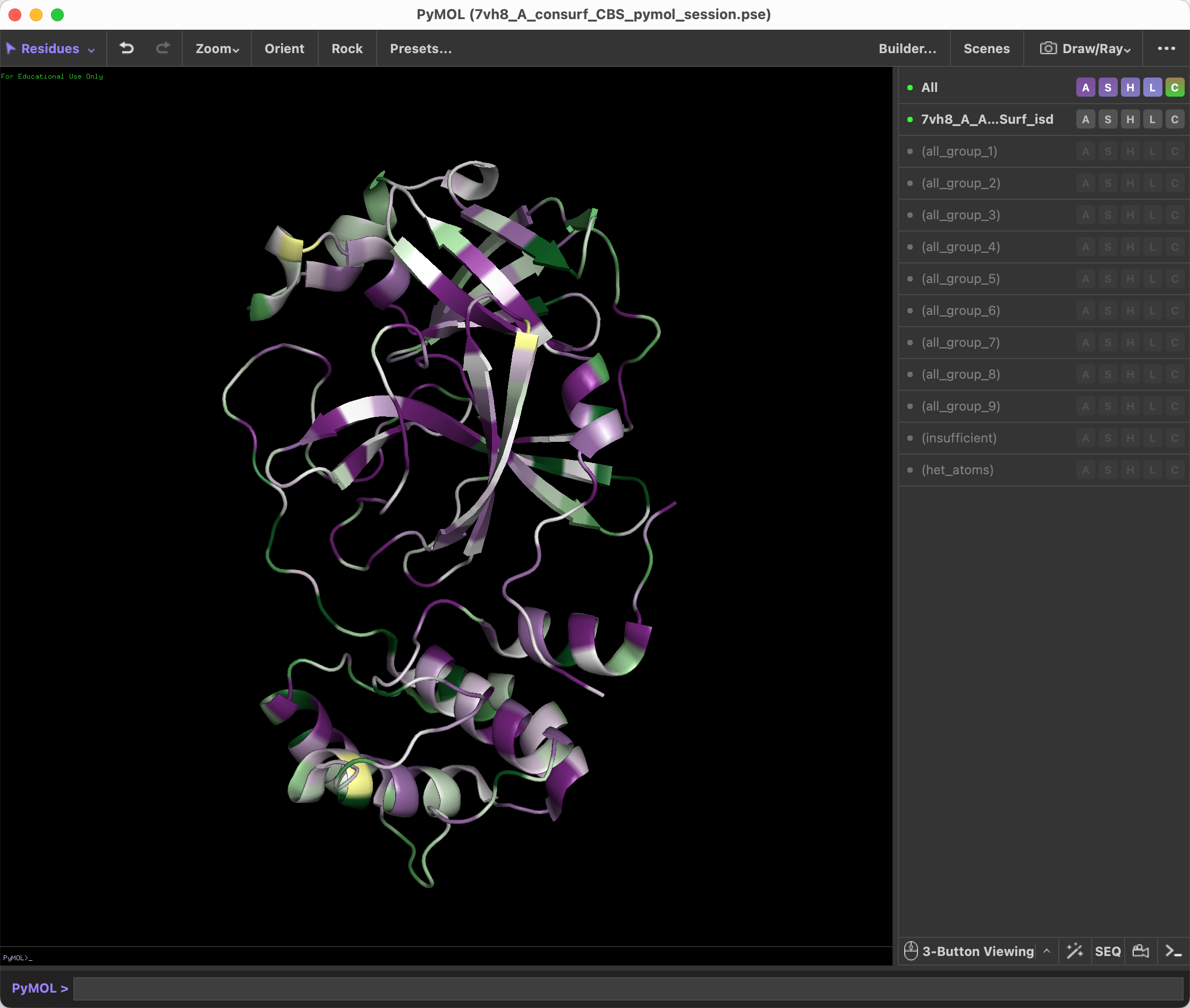
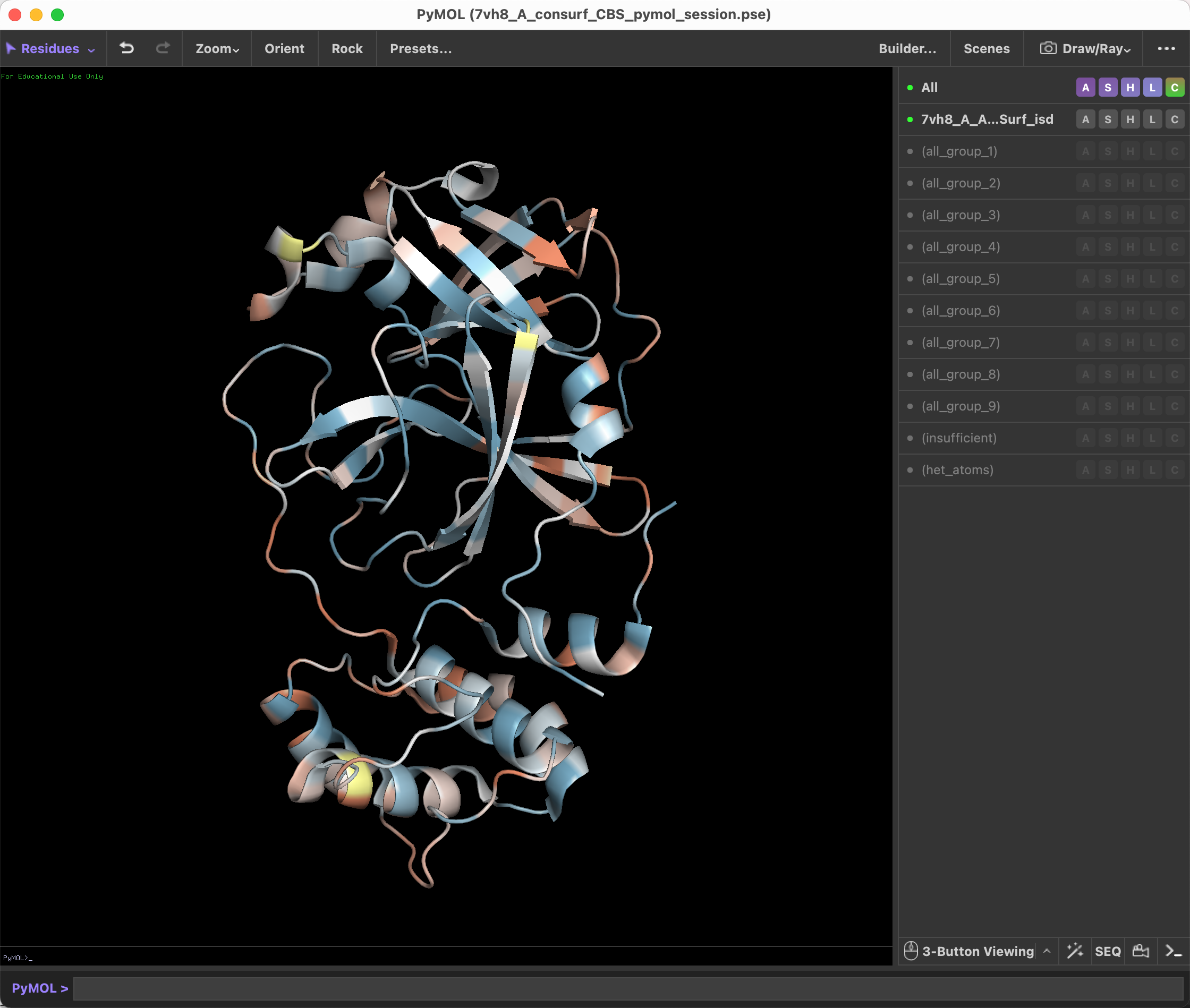
Load the ConSurf result in Pymol, and run the script from 'File -> Run Script...'.
:::grid {cols=2,rows=1,gap=12,type=images}
:::
此文由 Mix Space 同步更新至 xLog
原始链接为 https://xxu.do/posts/structure/Color-the-model-by-sequence-conservation-with-ConSurf